(Epi)genome replication: research topics
This team belongs to the UMR 9019 Genome Integrity and Cancers.
Our goal is to understand how cells copy both their DNA and epigenome during division. Disruptions in this process can lead to changes in cell identity and contribute to cancer.
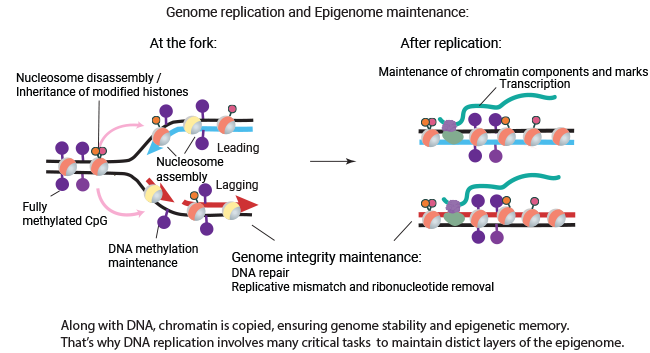
At each division of eukaryotic cells, the chromosomes must be precisely copied and transmitted from the mother cell to the daughter cells. Chromosome replication involves more than just the duplication of DNA; it also includes chromatin assembly, inheritance of epigenetic marks, maintenance of genome integrity, and seamless resumption of all genomic functions after replication. Our goal is to understand how mammalian cells link all these different processes to ensure efficient and accurate chromosome replication. We are seeking to reveal the interplay between different epigenome maintenance pathways and DNA replication and their roles in genome stability and cell epigenetic memory. Any alterations in these processes can lead to changes in cellular identity and cancer.
Chromosome replication is complex and fascinating. DNA replication must efficiently replicate various chromatin states and genomic features. DNA replication and epigenome replication are tightly interconnected, with many factors playing dual roles in both processes. We believe that this intimate relationship ensures the accurate transmission of both genetic and epigenetic information during cell division. Furthermore, the leading and lagging strands are replicated differently, and epigenome maintenance mechanisms need to align with the DNA replication asymmetry, which influences both genome evolution and epigenetic regulation.
We are currently focused on two main research topics in the lab
- We are investigating the role of nuclear proteins and structures in regulating chromosome replication, with a particular emphasis on the interaction between genome replication and chromatin's association with the nuclear lamina. Our research aims to uncover how alterations in the nuclear lamina affect genome replication dynamics, the maintenance of heterochromatin, and DNA damage response.
- We are exploring the connection between DNA replication and DNA methylation. Our goal is to understand the molecular mechanisms linking DNA replication with DNA methylation maintenance process, and how the dynamics of replication forks regulate the epigenome and influence cell fate.
Technologies
Our lab primarily utilizes and develops advanced genomic techniques, along with methods in molecular biology, genome editing, degron systems, and imaging in mouse and human cell lines. Additionally, we use chromatin assays and proteomics to investigate proteins involved at DNA replication forks. We are also advancing nanopore sequencing methods for our research.
Our specialized methods include:
OK-seq (Okazaki fragment sequencing): This technique is used to map DNA replication forks, measure replication fork dynamics, and quantify replication initiation and termination events genome-wide across a broad range of model organisms. We use OK-seq to study the molecular mechanisms of lagging strand processing in mammalian cells.
SCAR-seq (Sister Chromatids After Replication): This method allows us to map proteins bound to replication forks. Using SCAR-seq, we investigate how chromatin proteins and modifications—including both histone and non-histone factors—interact with DNA replication forks and replicated sister chromatids across the genome. These tools are critical for uncovering the mechanisms involved in epigenome maintenance, genome integrity, and replication dynamics in proliferating cells.
We are happy to share our expertise and open to new collaborations.
Collaborations
Constance Alabert (University of Dundee, UK, “Molecular Cell and Developmental Biology”)
Rodrigo Villaseñor (Ludwig Maximilian University of Munich, “Biomedical Center Munich Molecular Biology” )
Jean-Michel Arbona (ENS-Lyon, CNRS – INSERM-“Laboratory of Biology and Modeling of the Cell”)
Pierre-Antoine Defossez (University of Paris-Cité – CNRS – “Epigenetics and cell fate”
Magali Hennion (University of Paris – Cité CNRS “Epigenetics and cell fate”
Sophie Polo (University of Paris-Cité – CNRS – “Epigenetics and cell fate”
Funding
Our laboratory is funded by ATIP-Avenir, ANR, Gustave Roussy Foundation and EUGLOH.