Cell Biology of Organelle Networks: Towards a high resolution map of the cancer cell
This group belongs to the UMR 1279 - Tumor cell dynamics

The key feature of cells found in our body is the presence of a variety of different organelles. Organelles compartmentalize biochemical reactions that are otherwise incompatible. Moreover, organelles allow the existence of high local concentrations and therefore are intracellular platforms for signaling molecules. They support all fundamental processes including cell division, differentiation, proliferation, morphogenesis and cell motility. Yet, very little is known what happens to organelles during cancer: do they change in morphology, cellular positioning or function? Different to the genome, epigenome or proteomic atlases that have been established in recent years, no atlas about the organelle-level changes in cancer is presently available.
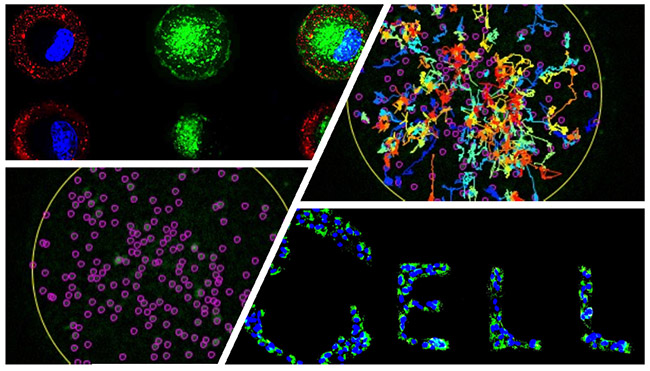
Our team investigates the intracellular landscape of organelles in the bladder cancer model. We combine bioengineering approaches for controlled cell culture condition with quantitative imaging to investigate which and how organelles change during cancer progression, drug treatment and resistance.

We have obtained a first quantitative description of the normal and pathologic landscape of the lysosomal compartment in a cellular model of bladder cancer, comparing normal human urothelium cells with bladder cancer cells of different stages. More than being the ‘stomach’ of eukaryotic cells, late endosomes/lysosomes have emerged as the cellular regulatory hub for metabolism and signaling, and thus, are important organelles for cell homeostasis. Our analysis identifies major changes in the positioning of lysosomes in high-grade bladder cancer cells that is accompanied by alterations in metabolic signaling pathways, occurring from lysosomes, and the molecular composition of this compartment.

Future studies will focus on organelle changes in samples from bladder cancer patients. Important axes are to reveal the individual molecular mechanisms associated with the metabolic reprogramming of bladder cancers, the immune suppression capabilities and resistance to current treatments. This will allow to potentially identify molecular predictors and to develop targeted therapies. Moreover, we aim at the discovery of individual cell vulnerabilities that will accelerate new customized cancer treatments. Our goal is to bridge a fundamental research program with concrete medical applications to improved patient diagnosis, treatment and survival.
Research topics
The team’s research topics are:
- mTORC1-TFEB nutrient signaling pathway in cancer cell metabolism
- Regulation of phosphoinositides (PI) and the PI3K pathway in cancer progression and resistance
- Secretion from lysosomes and cell migration
- Bioengineering approaches for extracellular matrix (ECM) for clinical applications
- Trafficking of cell surface receptors for targeted therapy