The team Biology of pediatric leukemia is part of the UMR 1170 Molecular dynamics of hematopoietic transformation.
The team is part of the childhood leukemia network CONECT-AML, the Paris Kid Cancer, the Institut Carnot OPALE, the Fédération d’Hématologie de l’Université Paris Saclay and coordinates the national PEDIAC program.
Pediatric cancers affect about 1 in 600 children and represent the second leading cause of death in children in France. The clinical characteristics of pediatric cancers suggest that they have a different molecular basis compared to similar cancers in adults. While recurrent genetic alterations have been identified in the past years, functional analyses are now required to identify how a normal cell become leukemic, why some alterations are specific for childhood and what are the mechanisms driving the leukemic cells that can be targeted for novel therapeutic perspectives.
Hematological malignancies represent 45% of pediatric cancers. The hematopoietic system is generally described as a hierarchy presenting, at its top, hematopoietic stem cells (HSC) that possess a unique capacity to transition from quiescence to long-term self-renewal potential in addition to the capacity to generate various proliferative progenitors that can ultimately differentiate toward the mature hematopoietic lineages. Acute myeloid leukemia (AML) is characterized by an accumulation of abnormal hematopoietic progenitors blocked in differentiation. They present recurrent genetic alterations including fusion oncogenes.
The main axes of our studies are to:
- 1-Study the crosstalk between stress response pathways and metabolic plasticity in normal and malignant hematopoiesis.
The goal is to decipher stress response of healthy and leukemic stem cells to uncover new metabolic vulnerabilities to target leukemic cells while sparing normal cells. - 2-Characterize the cellular and molecular trajectories during leukemia development.
The goal is to identify what are the mechanisms behind leukemia, how a normal cell becomes leukemia and why some alterations are specifically diagnosed during childhood. - 3-Identify functional dependencies and perform preclinical analyses.
The goal is to functionally identify mechanisms that represent specific vulnerabilities of leukemic cells and to test the efficacy of novel targeting strategies in preclinical settings using precise models of the disease.
Crosstalk between stress response pathways and metabolic plasticity in normal and malignant hematopoiesis
(PI: Marie-Laure Arcangeli)
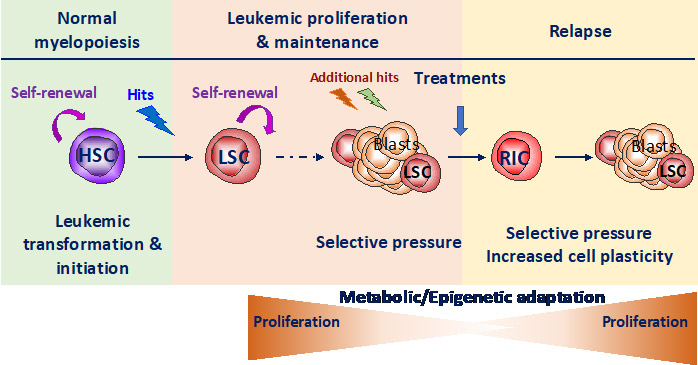
Our group is focused on the study of the crosstalk between stress response pathways, metabolic and epigenetic plasticity in normal and malignant hematopoiesis. HSC are often the cell of origin of the leukemia. Remission is frequently followed by relapse, mainly from resistant leukemic stem cells (LSC) that have adapted to treatment-induced stress by co-opting HSC functional properties. Therefore, there is a need to elucidate concurrently how both HSC and LSC adapt to stress (balance between quiescence and self-renewal) that relies largely on epigenetic and metabolic programs. Therefore, we aim to decipher stress response of healthy (HSC) and leukemic (LSC) stem cells to uncover new metabolic vulnerabilities to target LSC while sparing HSC.
Observations indicate that, once activated, HSC switch on stress response pathways and adapt to stress in particular by modulating metabolic pathways that leads to the modification of proteins encoded by epigenetic genes. Stress induces dramatic changes in transcriptional program in HSC with a switch in a signature from stem cell toward progenitor and a modification of the transcriptional pattern of epigenetic regulators (e.g. DNMT3a, TET2/3, IDH1/2 and ASXL1, Henry et al Stem cell Transl Med 2023). The control of oxidative stress response allows partial but essential protection of HSC fundamental properties (Henry et al Stem cell Transl Med 2023 et Henry et al Haematologica 2020, Exp Hematol 2020). Finally, we show that the stress response protein REDD1 participates to HSC protection by balancing ROS levels (Barroca et al Leukemia 2022).
In parallel, REDD1 overexpression is associated to bad prognosis and treatment resistance in AML. There are increasing evidence suggesting that REDD1 could be the intermediary between upstream stress response pathways and downstream metabolic rewiring leading to epigenetic remodeling.
Thus, we first propose, in gene candidate approach, to study relationships between REDD1, the antioxidant program, metabolism upon stress in human and mouse HSC and myeloid leukemia cells (using either shRNA technologies, human cell lines, PDX models and Redd1-/- deficient mice). Moreover, we will decipher REDD1 function in treatment resistance. This part is funded by INCA PLBIO (2024-2026) and performed in collaboration with JE Sarry (CRCT/IRCM, Toulouse/Montpellier).
More generally, we will combine high throughput approaches on human AML cell lines and mouse primary cells (combined RNA Sequencing and screening with CRSIPR library targeting metabolism and/or epigenetics) to decipher which stress response pathways are switched on and which epigenetic and metabolic pathway are remodeled. Indeed, we aim to uncover metabolic vulnerabilities in LSC that restore or provide sensitization to common treatments.
Cellular and molecular trajectories during leukemia development
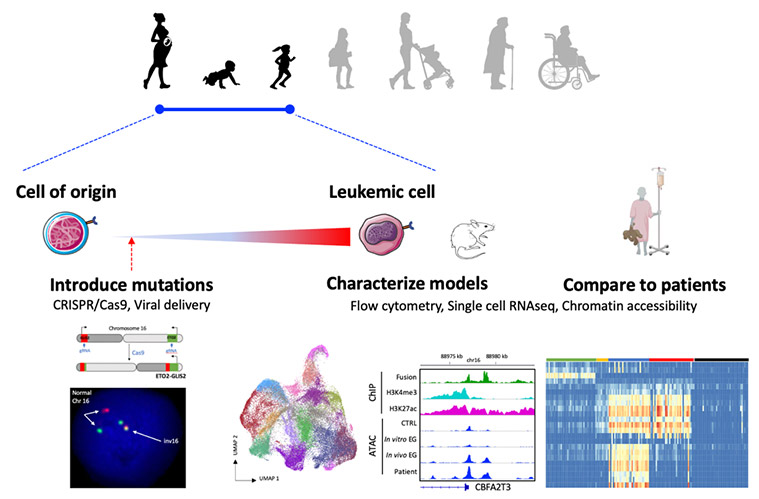
We characterize the genetic and epigenetic alterations found in leukemia and develop models to understand the functional contribution of each alteration as well as the stage of development of the cell of origin to leukemia initiation.
We have focused our work on pediatric acute megakaryoblastic leukemia (AMKL) and erythroleukemia (AEL) both generally associated with poor response to treatments and unfavorable prognosis.
Pediatric acute megakaryoblastic leukemia
We identified several recurrent fusion mutations and oncogenes involving regulators of gene expression (e.g. OTT-MAL and ETO2-GLIS2) (Mercher et al, PNAS 2001; Thiollier et al, JEM 2012). ETO2-GLIS2 is associated with the worst prognosis of pediatric de novo AMKL. We have shown that ETO2-GLIS2 binds DNA through both its ETO2 and GLIS2 parts, including at regulatory regions called "enhancers". It controls the expression of major transcription factors resulting in high ERG (essential for a stem cell-associated transcriptional program, including growth factor receptor KIT) and low GATA1 (a master regulator of erythro/megakaryocyte differentiation). In collaboration with C. Lobry and J. Chaumeil, we uncovered that the fusion controls the chromatin organization at the KIT/PDGFRA locus important for leukemia cell survival and proliferation (Benbarche et al. Science Adv 2022). Recently, we have observed that the fusion induces antagonistic effect on regulators of cell survival. Indeed, ETO2-GLIS2 induces activation of caspase 3 and cell death in naive cells while ETO2-GLIS2 leukemic cells present high expression of BCL2 (Aid et al. Leukemia 2023).
Acute erythroid leukemia (AEL)
We have developed with the group of Dr. J. Schwaller (Basel, Switzerland) a large collaborative work with teams from Gustave Roussy (Dr. S. DeBotton and Dr. J.B. Micol), French clinical centers (Dr. E. Delabesse: Toulouse, Dr. D. Birnbaum: Marseille, Dr. L. Garcon: Amiens), European centers (Dr. P. Vyas: England, Dr. E. Anguita: Spain, Dr. C. Dierks: Germany, Dr. A. Rambaldi: Italy, Dr. P. Valent: Austria) and international centers (Dr. M. Caroll: USA, Dr. J. Maciejewski: USA, Dr. S. Kazuya: Japan, Dr. C. Carmichael: Australia) to study human adult AEL. We identified genetic and transcriptional alterations and classified AEL patients into several molecular subgroups. Importantly, alterations of factors involved in GATA1 transcriptional complexes such as ERG, ETO2 or SKI, is found in at least 18% of the patients and functionally contribute to erythroid progenitor transformation in vitro and in mice (Fagnan et al Blood 2020).
Modeling leukemia initiation
We have developed a mouse model of doxycycline-inducible ETO2-GLIS2 expression (coll. with J. Schwaller, Basel, Switzerland) that 1-led to leukemia development upon induction of ETO2-GLIS2 expression, 2-showed that fetal cells are more permissive to transformation by the fusion than adult cells and 33-the phenotype of leukemia is associated with differential activity of several transcription factors, including GATA1 and CEBPA (Lopez et al. Cancer Discovery 2019).
We are now developing expression models of the ETO2-GLIS2 fusion starting from normal human cells through two approaches. First, we use induced pluripotent stem cells for their property to recapitulates early steps of human embryonic hematopoiesis. In a first model, we have shown that the fusion alters megakaryocyte differentiation, increases progenitor self-renewal, and recapitulates the transcriptional deregulations observed in patients but does not induce in vivo leukemia (Bertuccio et al Hemasphere 2020). To improve the model, we are currently performing precise engineering of normal iPSC using CRISPR/Cas9 to recreate the precise chromosomal alteration found in patient cells. We are also collaborating with the team of Dr. F. Pflumio (CEA, Fontenay-aux-roses) for the use of primary human cells at different stages of development. These models are studied using cellular and molecular technics, including single cell transcriptomes (scRNAseq), chromatin accessibility (ATAC-seq), mass spectrometry for protein interactions, multi-parameter flow cytometry (Aurora/Cytek).
Functional dependencies & preclinical analyses
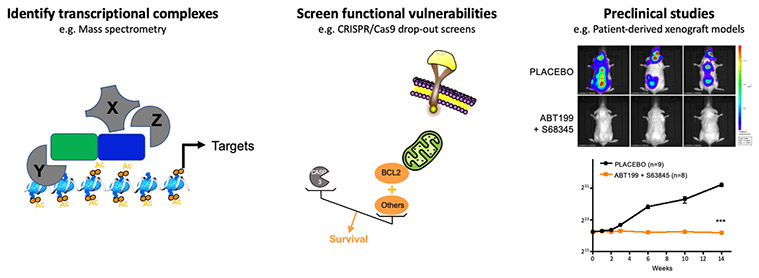
The characterization of transcriptional complexes altered by the fusion oncogenes or the identification of direct transcriptional targets allows to nominate candidates to be targeted by novel strategies in preclinical assays.
We have shown that the transcriptional program imposed by ETO2-GLIS2 depends on the functional interaction between ETO2-GLIS2 and ETO2 via the NHR2 domain. Indeed, ectopic expression of a peptide interfering with the NHR2 domain inhibits the expression of enhancer-associated genes, restores the expression balance of ERG and GATA1 factors, and abrogates the proliferation of ETO2-GLIS2 leukemia cells in in vivo models. These proof-of-principle data establish that a functional interference with the activity of transcriptional complexes involving ETO2-GLIS2 can inhibit proliferation/survival of leukemia cells (Thirant et al. Cancer Cell 2017, Lopez et al. Trends in Cancer 2017). In addition, as a follow-up of the work on erythroleukemia highlighting the role of ETO2 in transformation, ongoing studies are identifying some of the mechanisms controlling the positive role of ETO2 on transcriptional activation, including an important cofactor that can be target by small molecule inhibitor.
Emerging from the link between ETO2-GLIS2 and cell death regulation, a functional redundancy between BCL2 and MCL1, revealed in collaboration with P. Auberger (C3M, Nice), renders ETO2-GLIS2 cells virtually insensitive to inhibition by BCL2 or MCL1 inhibitors alone but highly sensitive to the combined inhibition, including in PDX models in vivo (Aid et al. Leukemia 2023). Following this work, we are collaborating with experts in pharmacology (Dr. F.X. Legrand, Université Paris Saclay) in order to overcome the current toxicities associated with combining these molecules.
As part of a collaboration led by E. Brunet (Imagine Institute), we are contributing to develop models of anaplastic large cell lymphoma (ALCL) with the NPM-ALK fusion. This work identified ROR2, as a novel surface marker uniquely expressed at the surface of NPM-ALK ALCL cells both in models and in patients (Babin et al iScience 2018 & Molecular Cancer 2022, Patent). This work provided a target for the development of a novel immunotherapy strategy in ALCL that acquire resistance to current therapies.
