Inflammation and cancer plasticity
This team belongs to the UMR 981 - Molecular predictors and new targets in oncology
Over a lifetime, our tissues accumulate cancer-related mutations, but not all of them lead to cancer. Our lab studies the mechanisms promoting some mutated cells to turn cancerous in some people while others do not. We are especially interested in how obesity and metabolic conditions increase the risk of lung and pancreatic cancers, which often have a mutation in a gene called KRAS, which drives cancer progression.
Research topics
Oncogenic mutations responsible for cancer accumulate in tissues over time, but most do not lead to a malignant tumour. The Inflammation and Cancer Plasticity Lab is part of the U981 Inserm unit and the IHU Prism at Gustave Roussy. Our lab seeks to understand the molecular and cellular mechanisms that drive the transformation of cells carrying an oncogenic mutation.
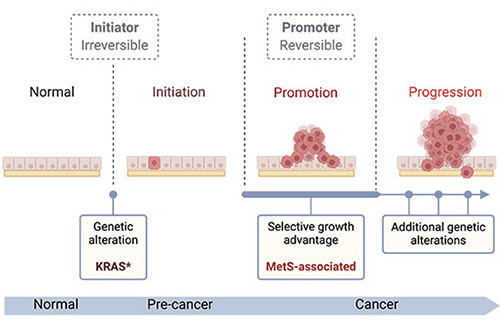
We are particularly interested in cancers with frequent KRAS mutations in the context of metabolic diseases associated with diet and obesity. KRAS, a member of the RAS oncogene family, is frequently mutated in cancer, leading to continuous activation of downstream signalling pathways that increase cell proliferation, promote motility, and inhibit cell death. KRAS activating mutations occur in approximately 30% of lung adenocarcinomas (LUAD) and 90% of pancreatic ductal adenocarcinomas (PDAC). Lung cancer remains the leading cause of cancer-related death in the population, and PDAC has the lowest survival rate of all major cancers, with incidence rates rising in younger populations (under 50).
Both cancers have been linked to obesity and metabolic syndrome (MetS)—a cluster of metabolic abnormalities diagnosed when individuals present with at least three of these criteria: central obesity, hypertension, hyperglycaemia, and dyslipidaemia (high triglycerides or low HDL cholesterol). Obesity and MetS are both increasing in the general population and in children, underlining the urgent need to understand how these conditions drive cancer initiation and progression.
Our research spans several areas of cancer biology and their interplay:
- Cancer Cell Plasticity – Understanding how metabolic diseases influence transcriptional heterogeneity within the pool of cancer cells, particularly their effect on cancer stem cells (CSCs), which are responsible for cancer progression and relapse after treatment.
- Tumour Microenvironment (TME) – Exploring the interplay between cancer and immune cells and understand how metabolic diseases influence this to shape tumour evolution.
- Systemic Drivers of Tumour Evolution – Identifying the systemics factors that promote KRAS-driven cancers in people with metabolic diseases and understanding how they drive cancer evolution.
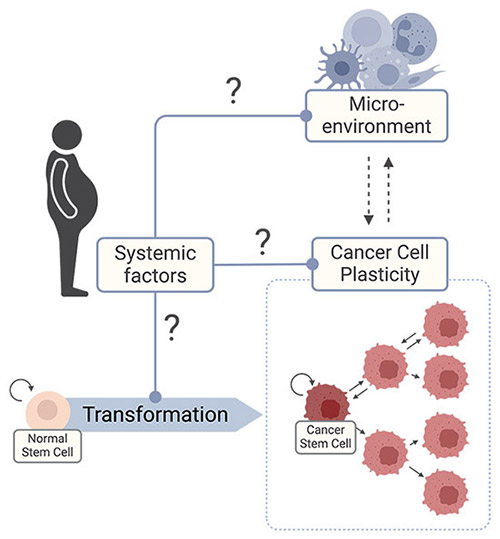
To address these questions, we integrate patient-derived samples, preclinical models, and advanced molecular profiling technologies:
- Patient-Derived Samples – Working with the Interception programme at Gustave Roussy, we access clinical samples from individuals at risk of developing cancer or with established malignancies. By collecting clinical data and paired samples from benign and malignant tumours within the same individuals, we investigate the presence of oncogenic mutations and the tumour microenvironment in both normal and tumoural tissues. These data are correlated with clinical and environmental factors to identify novel risk factors and elucidate their impact on cancer initiation and progression.
- Preclinical Models – We employ genetically engineered mouse models (GEMMs) with inducible oncogenic mutations and engineered tracking systems to study tumour initiation and progression under controlled conditions that mimic patient-specific risk factors. Additionally, we use organoid systems derived from human induced pluripotent stem cells (iPSCs) to model tumour evolution in controlled in vitro settings and characterise the molecular pathways involved.
- Advanced Molecular Profiling – We apply single-cell sequencing (e.g., CITE-Seq, scRNA-Seq) and spatial transcriptomics to dissect the cellular and molecular landscape of tumour initiation and progression at a single-cell resolution. These technologies provide unprecedented insights into the transcriptional and spatial heterogeneity of tumour and immune cell populations.
By deciphering the mechanisms underlying tumour initiation and progression in individuals at increased risk of cancer, our work aims to guide new prevention strategies, raise awareness of modifiable risk factors, and develop early therapeutic interventions for at-risk patients.
